The HPV genotype indicates risk
as well as the potential presence of
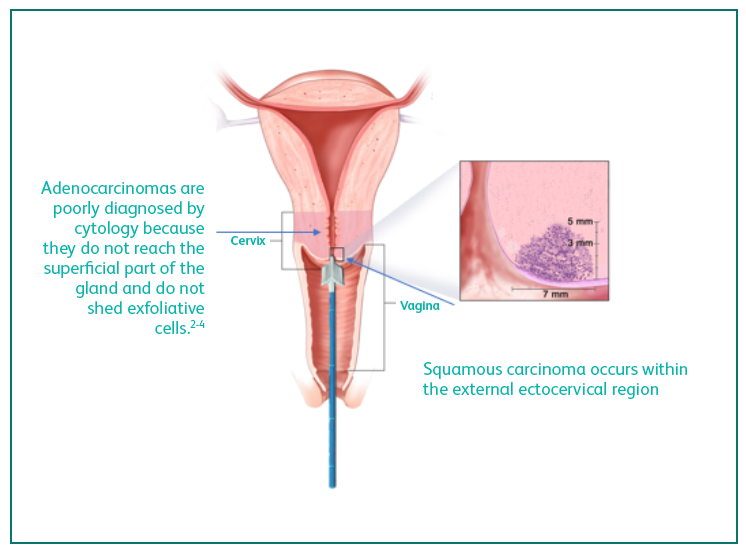
hard-to-detect adenocarcinomas
hard-to-detect adenocarcinomas

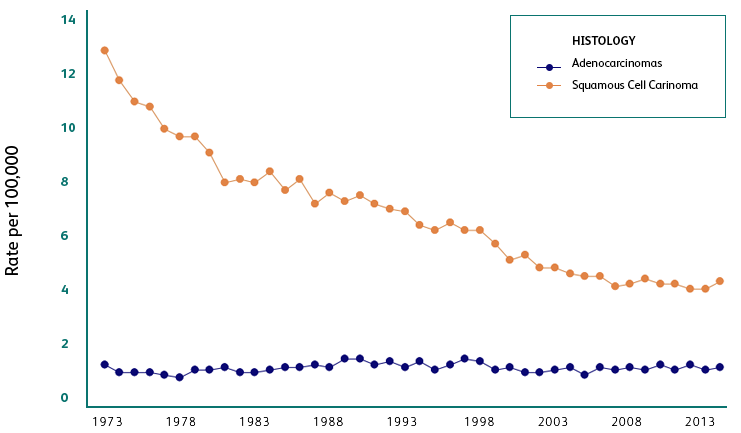
Adenocarcinomas are increasing in prevalence1 and more difficult to diagnose2-4 compared to squamous cell cancers.

HPV 16, 18 and 45 account for 94% of HPV (+) adenocarcinomas.5